
OCTOPUS v2.0: New improvements to help your plasmid sequencing scale
OCTOPUS v2.0: New improvements to help your plasmid sequencing scale
Right now, around the world, molecular biology labs like Octant's are hard at work engineering organisms. This often requires creating plasmids – circular strands of DNA – and using them to deliver to cells the instructions for new proteins. After creating plasmids, scientists usually verify them via DNA sequencing before investing additional time and resources in the transfection process. For verification, scientists typically use Sanger sequencing, a decades-old technique that (while tried and true) has missed out on years of exponential advances in DNA sequencing.
While Sanger sequencing has been adequate for most molecular biology labs, Octant, and other labs around the world, have been pioneering orders-of-magnitude improvements in the scale at which we are engineering plasmids and organisms. Our scale quickly outpaced the availability of reliable and fast Sanger sequencing services. Two years ago, we decided to engineer ourselves out of this bottleneck by designing a new plasmid verification system that would leverage the latest advances in DNA sequencing – OCTOPUS. Because we dream of a world where all scientists have easy access to cost-effective, scalable plasmid sequencing, we made this technology open source for anyone to use. We’ve come a long way in the two years since we first announced OCTOPUS, and want to share recent improvements to its capabilities and reflect on the impact this technology has had on Octant.
Why OCTOPUS
Before talking about OCTOPUS’s evolution at length, it’s worth summarizing why it’s so unique and some of its key features.
First, it just works. After running this pipeline once a week (sometimes more) for two years, we’ve rarely encountered more than a flipped plate or incorrect sample submission. The benchtop protocol and the computational pipeline are robust. Unlike Sanger sequencing based plasmid verification procedures, OCTOPUS leverages next-generation sequencing to vastly increase our throughput. Armed with this workflow, a research associate with just 1-2 weeks of training can process hundreds of samples a week and produce clean, whole-plasmid sequences for less than 10% of the total cost of a typical Sanger sequencing vendor.
Second, OCTOPUS’s simplicity drives greater scale. Because OCTOPUS is so easy, scientists collect whole-plasmid sequences more often during their projects. This means that we catch mutations, contaminations, and other unwanted mix-ups earlier and more often. With the burden of plasmid verification eased, scientists feel empowered to take on bolder cloning strategies. With OCTOPUS, a scientist assembling many complex plasmids in parallel can easily perform a “one-pot” reaction and screen as many products as they need.
OCTOPUS makes synthetic biology more delightful. Scientists at Octant don’t have to tediously design Sanger sequencing primers, patiently wait days (or, if you have too many samples, weeks) for a vendor to process their samples, or worry about checking traces and manually assembling contigs. They simply turn in their colonies and get back clean, whole-plasmid sequences 40 hours later.
We’ve found OCTOPUS so delightful that we want other scientists to enjoy it as well. At Octant, we believe it’s world versus disease, not us versus the world. As with Swab-Seq, we’ve made this technology open source so that other scientists can experience the delight of OCTOPUS cloning for themselves. In the past two years, OCTOPUS has matured from a scrappy startup workstream to an on-demand fixture of our synthetic biology stack. We again invite curious people to give this technology a try for themselves, because whether you're in a university sequencing core, a lean startup, or an established company– OCTOPUS can make a tremendous difference.

The Power of OCTOPUS v2.0
OCTOPUS began as a project between one computational scientist, one biologist, and an Octant apprentice. After mere weeks of development, they were sequencing hundreds of full-plasmids and were excited to share this initial protocol with the world. But in the past two years, we’ve also significantly improved OCTOPUS in several areas to further its throughput and usability:
- We switched to an RCA based protocol, making it dramatically easier to harvest DNA from bacteria and improving the overall operational efficiency and yield.
- We automated much of the protocol using the open-source Opentrons OT-2 liquid handling system and reduced the necessary hands-on time by more than 70%.
- We updated our documentation and improved our interfaces so that anyone, regardless of their computational background, can run the analysis pipeline.
- We published an actual Octant dataset for testing. You can download the raw sequencing data–right now, on your personal laptop–and try OCTOPUS for yourself.
Armed with these new improvements, it’s not uncommon for one lab worker to screen 1000+ plasmids weekly in two or three runs using about 25% of their time.
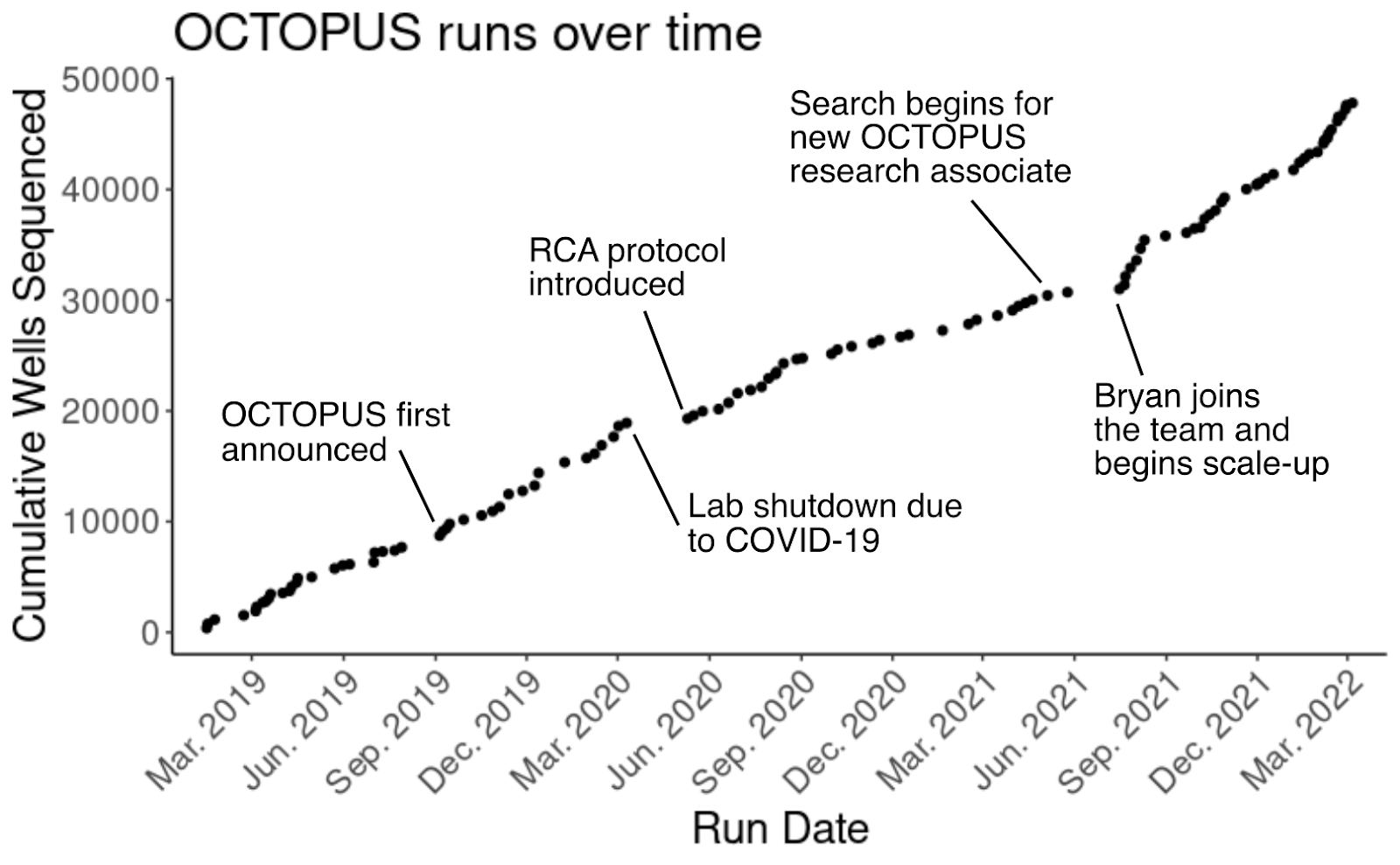
In the two years since the blog post was published, we’ve run this protocol over a hundred times and sequenced over 40,000 wells. By the time I joined Octant as an apprentice in August of 2020, sequencing plasmids on OCTOPUS was an Octant rite of passage. Everyone across the company – from our more technology-development-focused teams to our drug development biology teams–rely on this technology.
Where OCTOPUS is heading
Alongside four different kinds of coffee machines, multiplexed assays, and homebrewed kombucha on tap, OCTOPUS has become ingrained in Octant’s DNA. Our scientists rely on it for their most critical projects and new Octonauts learn it to produce high quality whole plasmid sequences within weeks of joining the team.
We will continue streamlining and optimizing its workflow, automating more of the protocol and further improving the ease and scale of the computational pipeline. We envision a completely automated procedure in which any scientist can walk up to the bench and run an entirely automated OCTOPUS (“AUTOPUS”), getting complete plasmid sequences in their inbox 40 hours later.
We’re proud of OCTOPUS’s capabilities, and there’s still room for improvement. In true open source spirit, please send us feedback (pull requests, patches, issues, etc.) to help us improve it. We hope you give it a try.

Will Owens

