
OCTOPUS 3.0 - Next-day turnaround of full plasmid sequencing directly from colonies
OCTOPUS 3.0 - Next-day turnaround of full plasmid sequencing directly from colonies
At Octant we’re deploying high-throughput biology and chemistry to engineer next-generation small molecule therapeutics that improve people’s lives. Our drug discovery platform, which we call the “Navigator,” searches for these drugs by screening compounds from our high-throughput SAR (HT-SAR) platform against custom cell lines that are engineered to report on biological activity (e.g., function, signaling, abundance, trafficking) using high-throughput synthetic biology.
Announcing OCTOPUS 3.0 – A Better Method for High-Throughput Full Plasmid Sequencing
Today we’re excited to share OCTOPUS 3.0, the latest edition of our in-house full plasmid sequencing platform that features two substantial upgrades:
* ONE-STEP library prep for easy execution and onboarding
* NEXT-DAY turnaround directly from colonies, faster than Sanger
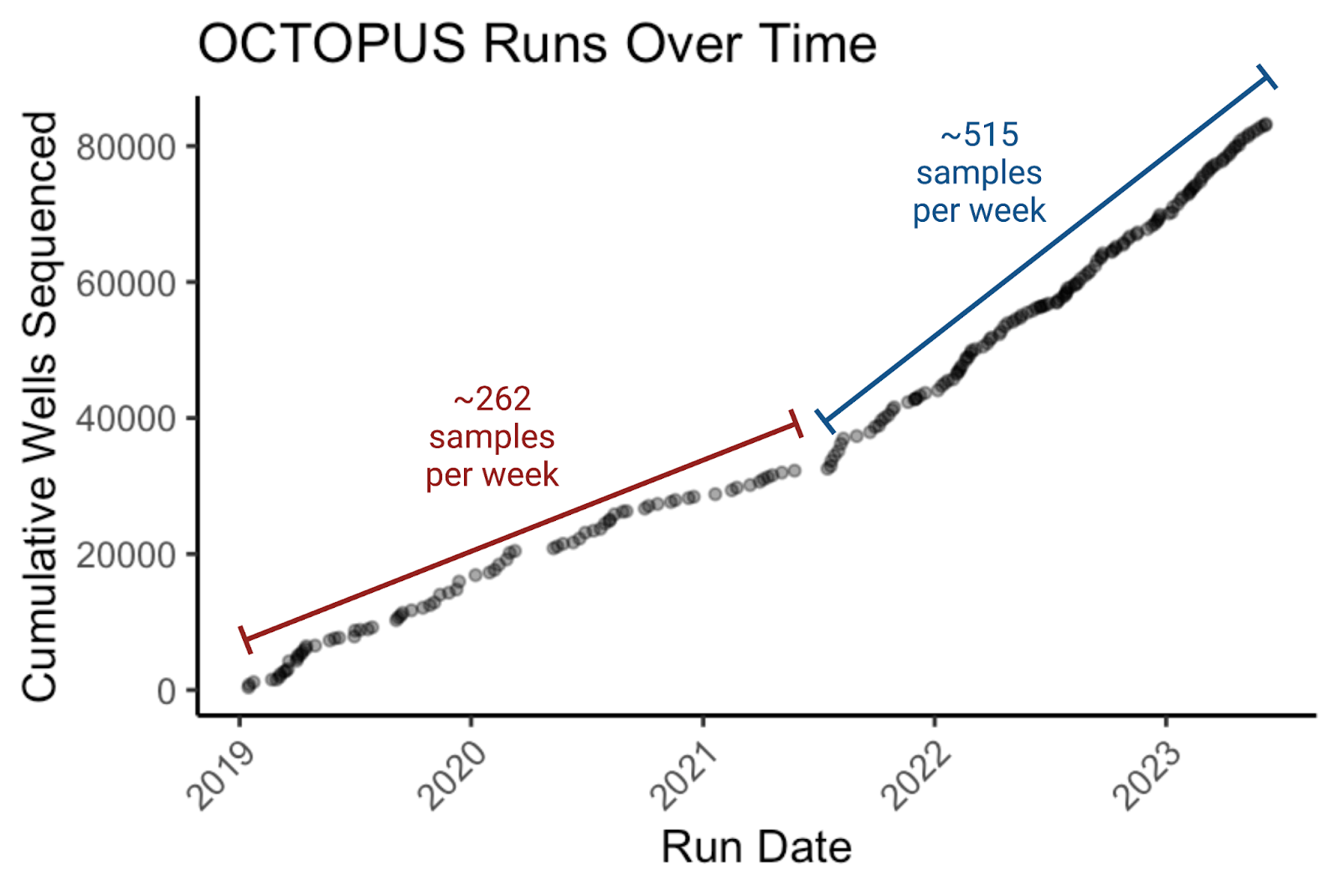
Engineering cell lines for high-throughput assay development requires extensive molecular cloning on the order of hundreds of plasmids a week. To ensure expected behavior in our cell models, all the upstream plasmid sequences have to be verified, which has been a bottleneck. Rather than outsource our sequencing, which would be expensive, slow, and throughput-limiting, we built an in-house high-throughput full-plasmid sequencing platform called OCTOPUS. OCTOPUS utilizes next-generation sequencing (NGS) to verify plasmids directly from bacterial colonies cheaply at-scale. While previous blog posts have focused on the origin of OCTOPUS and incorporating automation into the process, here we'll share the latest improvements to the NGS library prep and input generation that led us to achieve a dramatic reduction in turnaround time and the overhead required to run OCTOPUS.
Full Plasmid Sequencing Library Prep in One 90-minute Step
OCTOPUS library prep is now a single 90-minute step, making it 4X faster and easier than before! This protocol requires far fewer intermediate steps than OCTOPUS 2.0 by utilizing the ExpressPlex library prep kit, a new product created by seqWell. ExpressPlex collapses the traditionally separate NGS library prep steps of random primer extension (or tagmentation), bead capture, and PCR into a single one-hour thermocycling step, turning a long involved process into a fast painless one. This means OCTOPUS no longer requires veteran operator time, dramatically reducing overhead and turnaround and increasing accessibility.

No More Long Incubations for DNA Input Generation
OCTOPUS 3.0 reduces rolling circle amplification (RCA) from 8 hours to 3 hours! We’ve swapped out the standard phi29 polymerase for a more processive engineered form capable of reaching saturation within 3 hours. Reaching saturation for DNA input generation is critical because it guarantees sample-independent, quantitation-free normalization across every well. We’ve also put a spin on the RCA protocol that enriches the plasmid DNA of interest over contaminating DNA from the host transformant, leading to more coverage of the relevant data.

Next-Day Turnaround Enables Faster Iteration
These two changes culminate in shaving an entire day off OCTOPUS! With OCTOPUS 3.0, colonies picked this morning have their plasmids fully sequenced 24 hours later, a faster turnaround than Sanger service from the same input! For us, this often means the difference between starting an experiment that same week vs. waiting until the next for mission critical data.
OCTOPUS continues to be a game changer
Since its inception, OCTOPUS has fundamentally changed how we approach molecular and synthetic biology. The ease and scale with which we can sequence verify full plasmids directly from colonies, coupled with the low cost (about the price of a single Sanger reaction per sample) means that we can take riskier but more rewarding approaches to molecular cloning. Conservative estimates suggest OCTOPUS has saved Octant at least a million dollars in Sanger costs alone, not to mention the cost and effort required to order and submit Sanger sequencing primers. Moreover, the automated analysis pipeline that interprets sequencing data, eliminates the need for scientists to qualitatively analyze individual trace files. These massive savings and short turnaround times make OCTOPUS a cornerstone of the synthetic biology work at Octant and we’re excited to continue to share it with the community! With the simplicity of the current protocol, we’re confident no more barriers to entry exist. OCTOPUS is open-source, so we encourage others to try it, bolster our protocol and share their progress!
Try It Out Yourself & Learn More
See how you can streamline your in-house sequencing by adapting our protocol– check out our GitHub here!
To hear our Synthetic Biology Lead, Henry Chan, and Research Associate, Bryan Jiang, dive deeper into the evolution and application of OCTOPUS and answer some protocol questions, check out this joint webinar!

Bryan Jiang

